Building an Intake-esm catalog from CESM2 History Files#
This example covers how to build an intake-esm catalog from Community Earth System Model v2 (CESM2) model output. In this case, we use model output using the default component-set (compset) detailed in the CESM Quickstart Guide.
What’s a “history” file?#
A history file is the default output from CESM, where each file is a single time “slice” with every variable from the component of interest. These types of files can be difficult to work with, since often times one is interested in a time series of a single variable. Building a catalog can be helpful in accessing your data, querying for certain variables, and potentially creating timeseries files later down the road.
Let’s get started!
Imports#
The only parts of ecgtools we need are the Builder object and the parse_cesm_history parser from the CESM parsers! We import glob to take a look at the files we are parsing.
import glob
from ecgtools import Builder
from ecgtools.parsers.cesm import parse_cesm_history
Understanding the Directory Structure#
The first step to setting up the Builder object is determining where your files are stored. As mentioned previously, we have a sample dataset of CESM2 model output, which is stored in /glade/work/mgrover/cesm_test_data/
Taking a look at that directory, we see that there is a single case b.e20.B1850.f19_g17.test
glob.glob('/glade/work/mgrover/cesm_test_data/*')
['/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test']
Once we go into that directory, we see all the different components, including the atmosphere (atm), ocean (ocn), and land (lnd)!
glob.glob('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/*')
['/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/logs',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/cpl',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/lnd',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/esp',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/glc',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/rof',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/rest',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/wav',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ice']
If we go one step further, we notice that within each component, is a hist directory which contains the model output
glob.glob('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm/*/*.nc')[0:3]
['/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm/hist/b.e20.B1850.f19_g17.test.cam.h0.0002-08.nc',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm/hist/b.e20.B1850.f19_g17.test.cam.h0.0001-09.nc',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm/hist/b.e20.B1850.f19_g17.test.cam.h0.0002-07.nc']
If we take a look at the ocn component though, we notice that there are a few timeseries files in there…
glob.glob('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn/*/*.nc')[0:3]
['/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn/tseries/b.e20.B1850.f19_g17.test.pop.h.pCO2SURF.000101-001012.nc',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn/tseries/b.e20.B1850.f19_g17.test.pop.h.SiO3_RIV_FLUX.000101-001012.nc',
'/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn/tseries/b.e20.B1850.f19_g17.test.pop.h.graze_sp_zootot.000101-001012.nc']
When we setup our catalog builder, we will need to specify not including the timeseries (tseries) and restart (rest) directories!
Now that we understand the directory structure, let’s make the catalog.
Build the catalog!#
Let’s start by inspecting the builder object
?Builder
Init signature:
Builder(
root_path: pydantic.types.DirectoryPath,
extension: str = '.nc',
depth: int = 0,
exclude_patterns: List[str] = None,
njobs: int = -1,
) -> None
Docstring:
Generates a catalog from a list of files.
Parameters
----------
root_path : str
Path of root directory.
extension : str, optional
File extension, by default None. If None, the builder will look for files with
"*.nc" extension.
depth : int, optional
Recursion depth. Recursively crawl `root_path` up to a specified depth, by default 0
exclude_patterns : list, optional
Directory, file patterns to exclude during catalog generation.
These could be substring or regular expressions. by default None
njobs : int, optional
The maximum number of concurrently running jobs,
by default -1 meaning all CPUs are used.
File: /glade/work/mgrover/git_repos/ecgtools/ecgtools/builder.py
Type: type
Subclasses:
b = Builder(
# Directory with the output
"/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/",
# Depth of 1 since we are sending it to the case output directory
depth=1,
# Exclude the timeseries and restart directories
exclude_patterns=["*/tseries/*", "*/rest/*"],
# Number of jobs to execute - should be equal to # threads you are using
njobs=5,
)
Double check the object is set up…
b
Builder(root_path=PosixPath('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test'), extension='.nc', depth=1, exclude_patterns=['*/tseries/*', '*/rest/*'], njobs=5)
We are good to go! Let’s build the catalog by calling .build() on the object! By default, it will use the LokyBackend which is described in the Joblib documentation.
We also add in the parser here! By default, the parsers use a default_streams dictionary formatted as follows to parse the files:
This dictionary follows the convention:
{'stream': {'component': 'some_component', 'frequency': 'frequency_num'}
Here is an example of the first few!
{'cam.h0': {'component': 'atm', 'frequency': 'month_1'},
'cam.h1': {'component': 'atm', 'frequency': 'day_1'},
}
b = b.build( # Use the parse_cesm_history parsing function
parse_cesm_history,
)
[Parallel(n_jobs=5)]: Using backend LokyBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 6 out of 12 | elapsed: 0.2s remaining: 0.2s
[Parallel(n_jobs=5)]: Done 9 out of 12 | elapsed: 0.2s remaining: 0.1s
[Parallel(n_jobs=5)]: Done 12 out of 12 | elapsed: 0.2s remaining: 0.0s
[Parallel(n_jobs=5)]: Done 12 out of 12 | elapsed: 0.2s finished
[Parallel(n_jobs=5)]: Using backend LokyBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 8 tasks | elapsed: 1.5s
[Parallel(n_jobs=5)]: Done 62 tasks | elapsed: 5.1s
[Parallel(n_jobs=5)]: Done 152 tasks | elapsed: 12.3s
[Parallel(n_jobs=5)]: Done 264 out of 264 | elapsed: 16.8s finished
/glade/work/mgrover/git_repos/ecgtools/ecgtools/builder.py:180: UserWarning: Unable to parse 5 assets/files. A list of these assets can be found in `.invalid_assets` attribute.
parsing_func, parsing_func_kwargs
Inspect the Catalog#
Now that the catalog is built, we can inspect the dataframe which is used to create the catalog by calling .df on the builder object
b.df
| component | stream | case | date | frequency | variables | path | |
|---|---|---|---|---|---|---|---|
| 0 | atm | cam.h0 | b.e20.B1850.f19_g17.test | 0002-08 | month_1 | [date, datesec, date_written, time_written, nd... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 1 | atm | cam.h0 | b.e20.B1850.f19_g17.test | 0001-09 | month_1 | [date, datesec, date_written, time_written, nd... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 2 | atm | cam.h0 | b.e20.B1850.f19_g17.test | 0002-07 | month_1 | [date, datesec, date_written, time_written, nd... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 3 | atm | cam.h0 | b.e20.B1850.f19_g17.test | 0003-05 | month_1 | [date, datesec, date_written, time_written, nd... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 4 | atm | cam.h0 | b.e20.B1850.f19_g17.test | 0002-01 | month_1 | [date, datesec, date_written, time_written, nd... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 259 | ice | cice.h | b.e20.B1850.f19_g17.test | 0001-08 | month_1 | [hi, hs, snowfrac, Tsfc, aice, uvel, vvel, uat... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 260 | ice | cice.h | b.e20.B1850.f19_g17.test | 0001-03 | month_1 | [hi, hs, snowfrac, Tsfc, aice, uvel, vvel, uat... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 261 | ice | cice.h | b.e20.B1850.f19_g17.test | 0002-11 | month_1 | [hi, hs, snowfrac, Tsfc, aice, uvel, vvel, uat... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 262 | ice | cice.h | b.e20.B1850.f19_g17.test | 0002-10 | month_1 | [hi, hs, snowfrac, Tsfc, aice, uvel, vvel, uat... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
| 263 | ice | cice.h | b.e20.B1850.f19_g17.test | 0003-12 | month_1 | [hi, hs, snowfrac, Tsfc, aice, uvel, vvel, uat... | /glade/work/mgrover/cesm_test_data/b.e20.B1850... |
259 rows × 7 columns
The resultant dataframe includes the:
Component
Stream
Case
Date
Frequency
Variables
Path
We can also check to see which files were not parsed by calling .invalid_assets
b.invalid_assets
| INVALID_ASSET | TRACEBACK | |
|---|---|---|
| 15 | /glade/work/mgrover/cesm_test_data/b.e20.B1850... | Traceback (most recent call last):\n File "/g... |
| 28 | /glade/work/mgrover/cesm_test_data/b.e20.B1850... | Traceback (most recent call last):\n File "/g... |
| 34 | /glade/work/mgrover/cesm_test_data/b.e20.B1850... | Traceback (most recent call last):\n File "/g... |
| 130 | /glade/work/mgrover/cesm_test_data/b.e20.B1850... | Traceback (most recent call last):\n File "/g... |
| 191 | /glade/work/mgrover/cesm_test_data/b.e20.B1850... | Traceback (most recent call last):\n File "/g... |
b.invalid_assets.INVALID_ASSET.values[0]
PosixPath('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/atm/hist/b.e20.B1850.f19_g17.test.cam.i.0002-01-01-00000.nc')
If you wanted to add a stream, say cam.i for example, which are instantaneous CAM files, you would use the parsing_func_kwargs argument within the build function!
First, we setup a variable to contain the new stream information.
new_streams = {'cam.i': {'component': 'atm', 'frequency': 'instantaneous'}}
b = b.build(parse_cesm_history, parsing_func_kwargs={'user_streams_dict': new_streams})
[Parallel(n_jobs=5)]: Using backend LokyBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 6 out of 12 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=5)]: Done 9 out of 12 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=5)]: Done 12 out of 12 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=5)]: Done 12 out of 12 | elapsed: 0.0s finished
[Parallel(n_jobs=5)]: Using backend LokyBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 8 tasks | elapsed: 0.6s
[Parallel(n_jobs=5)]: Done 62 tasks | elapsed: 4.3s
[Parallel(n_jobs=5)]: Done 152 tasks | elapsed: 11.1s
[Parallel(n_jobs=5)]: Done 264 out of 264 | elapsed: 15.5s finished
/glade/work/mgrover/git_repos/ecgtools/ecgtools/builder.py:180: UserWarning: Unable to parse 2 assets/files. A list of these assets can be found in `.invalid_assets` attribute.
parsing_func, parsing_func_kwargs
b.invalid_assets.INVALID_ASSET.values[0]
PosixPath('/glade/work/mgrover/cesm_test_data/b.e20.B1850.f19_g17.test/ocn/hist/b.e20.B1850.f19_g17.test.pop.hv.nc')
It appears that one of the invalid assets is a pop.hv stream, which is a time-invariant dataset we would not neccessarily be interested in looking at. If there is a file you think should be included in the resultant catalog but isn’t, be sure to add it to the default_streams dictionary used in the parsing tool!
Save the Catalog#
b.save(
# File path - could save as .csv (uncompressed csv) or .csv.gz (compressed csv)
"/glade/work/mgrover/cesm-hist-test.csv",
# Column name including filepath
path_column_name='path',
# Column name including variables
variable_column_name='variables',
# Data file format - could be netcdf or zarr (in this case, netcdf)
data_format="netcdf",
# Which attributes to groupby when reading in variables using intake-esm
groupby_attrs=["component", "stream", "case"],
# Aggregations which are fed into xarray when reading in data using intake
aggregations=[
{
"type": "join_existing",
"attribute_name": "date",
"options": {"dim": "time", "coords": "minimal", "compat": "override"},
}
],
)
Saved catalog location: /glade/work/mgrover/cesm-hist-test.json and /glade/work/mgrover/cesm-hist-test.csv
/glade/u/home/mgrover/miniconda3/envs/cesm2-marbl/lib/python3.7/site-packages/ipykernel_launcher.py:17: UserWarning: Unable to parse 2 assets/files. A list of these assets can be found in /glade/work/mgrover/invalid_assets_cesm-hist-test.csv.
Using the Catalog#
You’ll notice the resultant filepaths are output when calling .save - which you could use within your intake-esm open_esm_datastore function.
Additional Imports#
# Import ast which helps with parsing the list of variables
import ast
# Import intake-esm
import intake
Use the catalog to read in data#
col = intake.open_esm_datastore(
"/glade/work/mgrover/cesm-hist-test.json",
csv_kwargs={"converters": {"variables": ast.literal_eval}},
sep="/",
)
col
None catalog with 10 dataset(s) from 262 asset(s):
| unique | |
|---|---|
| component | 6 |
| stream | 10 |
| case | 1 |
| date | 79 |
| frequency | 4 |
| variables | 1449 |
| path | 262 |
cat = col.search(
variables='TEMP',
)
cat
None catalog with 1 dataset(s) from 36 asset(s):
| unique | |
|---|---|
| component | 1 |
| stream | 1 |
| case | 1 |
| date | 36 |
| frequency | 1 |
| variables | 434 |
| path | 36 |
dsets = cat.to_dataset_dict(cdf_kwargs={'use_cftime': True, 'chunks': {'time': 10}})
--> The keys in the returned dictionary of datasets are constructed as follows:
'component/stream/case'
dsets
{'ocn/pop.h/b.e20.B1850.f19_g17.test': <xarray.Dataset>
Dimensions: (nlat: 384, nlon: 320, time: 36, z_t: 60)
Coordinates:
* time (time) object 0004-01-01 00:00:00 ... 0002-03-01 00:00:00
* z_t (z_t) float32 500.0 1.5e+03 2.5e+03 ... 5.125e+05 5.375e+05
ULONG (nlat, nlon) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
ULAT (nlat, nlon) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
TLONG (nlat, nlon) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
TLAT (nlat, nlon) float64 dask.array<chunksize=(384, 320), meta=np.ndarray>
Dimensions without coordinates: nlat, nlon
Data variables:
TEMP (time, z_t, nlat, nlon) float32 dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
Attributes:
contents: Diagnostic and Prognostic Variables
cell_methods: cell_methods = time: mean ==> the variable value...
time_period_freq: month_1
title: b.e20.B1850.f19_g17.test
intake_esm_varname: ['TEMP']
revision: $Id$
source: CCSM POP2, the CCSM Ocean Component
calendar: All years have exactly 365 days.
history: none
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/eaton/netcdf...
model_doi_url: https://doi.org/10.5065/D67H1H0V
intake_esm_dataset_key: ocn/pop.h/b.e20.B1850.f19_g17.test}
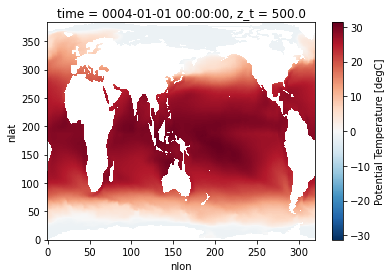
Let’s plot a quick figure from the dataset!
dsets['ocn/pop.h/b.e20.B1850.f19_g17.test'].TEMP.isel(time=0, z_t=0).plot();